CopyRight 2009-2020 © All Rights Reserved.版权所有: 中国海关未经授权禁止复制或建立镜像
菜豆晕疫病菌特异引物/探针检测效果评价
作者:牟桂萍 闫晓东 黎玮琪 易建平
牟桂萍 闫晓东 黎玮琪 易建平
牟桂萍 1,2 闫晓东 1,2 黎玮琪 1,2 易建平 3 *
摘 要 为了筛选菜豆晕疫病菌(Pseudomonas savastanoi pv. phaseolicola,Psph)最优特异引物/探针,本研究利用文献报道的14对Psph特异引物/探针,分别测试了Psph及近似种菌株59株,对其特异性和灵敏度进行测试和评价。特异性测试结果表明,常规PCR引物HM6/HM13、62a/63a和52B/8F特异性强,根据产物大小优先选择52B/8F;实时荧光PCR探针PsF-tox 286P和SSRP_P均有强特异性,但SSRP_P只能检测菜豆Psph,不能检测绿豆Psph。灵敏度测试结果表明,引物52B/8F的灵敏度为50 pg菌体DNA,探针PsF-tox 286P的灵敏度为500 fg菌体DNA。实际样品检测结果表明,引物52B/8F和探针PsF-tox 286P均可用于豆类样品Psph检测。
关键词 菜豆晕疫病菌;检测;引物;聚合酶链式反应
Evaluation of Specific Primers/Probes for Detecting Pseudomonas savastanoi pv. phaseolicola
MOU Gui-Ping 1,2 YAN Xiao-Dong 1,2 LI Wei-Qi 1,2 YI Jian-Ping 3*
Abstract In order to screen the optimal specific primers/probes of Pseudomonas savastanoi pv. phaseolicola (Psph), 59 strains of Psph and relative species were tested using 14 Psph-specific primers/probes reported in the literature. Their specificity and sensitivity were tested and evaluated. Specificity test results showed that conventional PCR primers HM6/HM13, 62a/63a and 52B/8F had strong specificity, and 52B/8F was preferred based on the product size. The real-time fluorescent PCR probes PsF-tox 286P and SSRP_P both exhibited strong specificity, but SSRP_P could only detect Psph in Phaseolus vulgaris L., not in Phaseolus radiatus L.. The sensitivity tests showed that the sensitivity of primer 52B/8F and probe PsF-tox 286P was 50 pg bacterial DNA and 500 fg bacterial DNA, respectively. The results of actual sample detection showed that both primer 52B/8F and probe PsF-tox 286P could be used for the detection of Psph in legume samples.
Keywords Pseudomonas savastanoi pv. phaseolicola (Psph); detection; primer; polymerase chain reaction (PCR)
菜豆晕疫病菌(Pseudomonas savastanoi pv. phaseolicola,Psph)是我国重要的进境植物检疫性有害生物,可引起菜豆和绿豆晕疫病。病菌寄主范围广泛,包括菜豆(Phaseolus vulgaris)、宽叶菜豆(P. acutifolius)、荷包豆(P. coccineus)、利马豆(P. lunatus)、木豆(Cajanus cajan)、距瓣豆(Centrosema sp.)、山蚂蝗(Desmodium spp.)、大豆(Glycine max)、扁豆(Lablab purpureus)、兵豆(Lens culinaris)、紫花大翼豆(Macroptilium atropurpureum)、多年生大豆(Neonotonia wightii)、豆薯(Pachyrhizus erosus)、豌豆(Pisum sativum)、葛藤(Pueraria lobata)、野葛(P. thunbergiana)、赤豆(Vigna angularis)、绿豆(V. radiata)、豇豆(V. unguiculata)等。
菜豆晕疫病菌主要通过带菌种子传播,在种子上可存活2年以上,0.02%的种子带菌率就可造成病害的流行。病菌主要为害叶片,也侵染豆荚和种子。带病种子长出的幼茎呈环状腐烂,表现为萎蔫并导致幼苗死亡。在气候凉爽潮湿的地区,病害可导致菜豆产量损失高达80%~100%,因此快速准确的分子生物学检测方法显得至关重要。目前对于Psph的分子生物学检测方法主要有常规聚合酶链式反应(Polymerase Chain Reaction,PCR)和实时荧光PCR方法。本文收集了文献报道的14对Psph特异引物/探针,对其特异性和灵敏度进行评价,筛选出最优引物/探针,并对实际样品进行应用测试。
1 材料与方法
1.1 引物/探针
共收集文献报道的14对Psph特异引物/探针(表1),引物/探针由生工生物工程(上海)股份有限公司合成。根据Psph基因序列,psp-JH-F、psp-JH-R、psp-JH-F-ne、psp-JH-R-ne 由原始文献的兼并引物调整为Psph特异引物。
1.2 材料
供试菌株:共收集菌株59株,其中包括25株Psph,菌株除来源于美国典型菌种保藏中心(American Type Culture Collection,ATCC)、比利时根特大学微生物实验室(Laboratorium Voor Microbiologie, Rijs Universiteit, Gent, Belgium,LMG)、英国植物病原细菌保藏中心(The National Collection of Plant Pathogenic Bacteria,NCPPB)外,中国检验检疫科学研究院、浙江大学等单位提供了部分试验菌株,其余为本实验室保存的菌株,供试菌株及其来源见表2。
营养琼脂(Nutrient Agar,NA)培养基:蛋白胨10 g、牛肉浸膏3 g、葡萄糖5 g、酵母提取物1 g、琼脂粉15 g、蒸馏水1 L,121℃ 20 min灭菌使用。
试剂:DNA提取试剂盒DP302(天根生化科技(北京)有限公司);DL2000 DNA marker(TaKaRa);2×Taq PCR Master Mix(TaKaRa);Premix Ex Taq (Probe qPCR) (TaKaRa);其余试剂均为分析纯。
样品:供试菜豆和绿豆种子样品共24个,由中国农业科学院作物科学研究所提供。
1.3 基因组DNA的提取
供试菌株在NA培养基上划线培养,挑取单菌落纯化后用无菌水洗脱,菌悬液采用细菌基因组DNA提取试剂盒提取DNA,用核酸测定仪测定DNA浓度和纯度,-20℃保存备用。
1.4 特异性测试
1.4.1 测试方法
采用14对特异性引物/探针对59株菌株的基因组DNA进行常规PCR或实时荧光PCR检测,以Psph菌株LMG2245的DNA作为阳性对照,以灭菌去离子水作空白对照,反应体系和反应程序参考各特异性引物/探针的原始文献。常规PCR扩增产物用1.5%琼脂糖凝胶1×TAE缓冲液电泳,溴化乙锭(Ethidium Bromide,EB)染色后用凝胶成像系统分析。
1.4.2 结果判定
常规PCR结果判定方法:阳性对照扩增出目标条带,空白对照无目标条带的前提下,待测菌株扩增条带与阳性对照大小一致,判定为阳性;待测菌株没有扩增条带或与阳性对照大小不一致,判定为阴性。
实时荧光PCR结果判定方法:阳性对照和空白对照均正常的前提下,待测菌株出现典型扩增曲线判定为阳性;无典型扩增曲线判定为阴性。
1.5 灵敏度测试
取Psph阳性菌株LMG2245的基因组DNA,经核酸蛋白测定仪测定浓度后进行梯度稀释,使DNA终浓度为50 ng/μL、5 ng/μL、500 pg/μL、50 pg/μL、5 pg/μL、500 fg/μL、50 fg/μL,分别用特异性测试筛选出的最优引物/探针进行PCR扩增,再进行灵敏度测试,反应体系和反应程序参考1.4.1。
1.6 样品检测
采用筛选出的最优引物/探针,对24个供试菜豆和绿豆样品进行PCR检测。选取3个经PCR检测Psph阳性样品表面消毒,浸泡过夜,浸泡液系列稀释后涂布NA平板,进行病原菌的分离培养。
2 结果与分析
2.1 特异性测试
采用14对特异性引物/探针对59株菌株的基因组DNA进行常规PCR或实时荧光PCR检测,测试结果见表2。
12对特异引物常规PCR测试结果显示,HM6/HM13(序号1)、62a/63a(序号5)和52B/8F(序号10)扩增25株Psph阳性菌株均扩增出现目的片段条带,且近似种均没有扩增条带,说明这3对引物可特异性地扩增Psph。以上3对引物的扩增片段大小分别为1900 bp、1000 bp和419 bp,由于HM6/HM13和62a/63a扩增产物片段较长,需要更长的PCR反应时间。为提高效率,缩短检测时长,优先选择52B/8F进行检测。其余9对引物对Psph近似种有强阳性或弱阳性扩增,表现出较差的特异性,分别是P5.1/P3.1(序号2)、P5.2/P3.2(序号3)、HB14F/HB14R(序号4)、PHA19/PHA95(序号6)、P3004L/P3004R(序号7)、psp-JH-F/psp-JH-R(序号8)、psp-JH-F-ne/psp-JH-R-ne(序号9)、24B/24F(序号11)、PSPF1/PSPR2(序号12)。值得注意的是,P3004L/P3004R(序号7)测试结果表明,有1株来自于菜豆的Psph阳性菌株和11株来自于绿豆的Psph阳性菌株出现目的片段条带,其余13株来自于菜豆的Psph阳性菌株无扩增条带。
2对特异引物探针法实时荧光PCR测试结果显示,探针PsF-tox 286P(序号13)扩增25株Psph阳性菌株均有扩增曲线,结果为阳性,其余34株近似种菌株均无扩增曲线,结果为阴性,说明探针特异性好。探针SSRP_P(序号14)测试结果表明,仅14株Psph阳性菌株有扩增曲线,结果为阳性,另11株Psph阳性菌株和近似种菌株均无扩增曲线,结果为阴性,说明探针易出现假阴性,只能检测部分Psph菌株,不能检测全部Psph菌株。因此,推荐探针PsF-tox 286P检测Psph。
2.2 灵敏度测试
Psph阳性菌株LMG2245的基因组DNA,梯度稀释至终浓度为50 ng/μL、5 ng/μL、500 pg/μL、50 pg/μL、5 pg/μL、500 fg/μL、50 fg/μL,分别测试引物52B/8F和探针PsF-tox 286P的灵敏度。结果表明,引物52B/8F常规PCR方法的检测灵敏度为50 pg菌体DNA(图1),探针PsF-tox 286P实时荧光PCR方法的检测灵敏度为500 fg 菌体DNA(图2)。探针PsF-tox 286P实时荧光PCR方法的检测灵敏度比引物52B/8F常规PCR方法的检测灵敏度高100倍。
2.3 实际样品检测
引物52B/8F和探针PsF-tox 286P分别检测24个菜豆和绿豆样品,样品编号及来源信息见表3。引物52B/8F检测样品结果表明,17个样品不同程度阳性(图3),探针PsF-tox 286P检测样品结果表明,23个样品不同程度阳性,样品Ct值从20.5~35.8不等,其中样品17-2中Psph阳性最强(表3)。将两种检测方法对比发现,实时荧光PCR Ct值越低,对应的样品常规PCR条带越亮,说明样品Psph阳性越强。6个实时荧光PCR Ct值>30的样品(样品编号15-1、23-1、20-2、22-2、27-2、28-2),常规PCR未出现目标条带,说明这些样品带菌量低,灵敏度较高的探针PsF-tox 286P实时荧光PCR方法可以检测到阳性,但灵敏度相对较低的引物52B/8F常规PCR方法检测不到阳性。将阳性最强的3个样品(10-1、17-2、24-2)表面消毒、浸泡,浸泡液系列稀释后涂布NA平板,得到的疑似菌落参考GB/T 28075—2011《 萨氏假单胞杆菌菜豆生致病型检疫鉴定方法》 和周密密等 报道的方法,经过菌落PCR、多序列分析、致病性测试,鉴定为Psph。
表3 实时荧光PCR探针PsF-tox 286P测试样品
Table 3 Sample test of real-time PCR assay using probe
PsF-tox 286P
样品编号 | 寄主 | Ct值 | 样品编号 | 寄主 | Ct值 |
10-1 | 绿豆 | 21.0 | 10-2 | 菜豆 | 24.8 |
11-1 | 绿豆 | 22.3 | 16-2 | 菜豆 | 22.5 |
12-1 | 绿豆 | 26.4 | 17-2 | 菜豆 | 20.5 |
13-1 | 绿豆 | 30.2 | 18-2 | 菜豆 | 24.2 |
14-1 | 绿豆 | 34.1 | 20-2 | 菜豆 | — |
15-1 | 绿豆 | 35.5 | 21-2 | 菜豆 | 32.1 |
16-1 | 绿豆 | 35.3 | 22-2 | 菜豆 | 31.1 |
17-1 | 绿豆 | 36.3 | 23-2 | 菜豆 | 32.0 |
18-1 | 绿豆 | 25.2 | 24-2 | 菜豆 | 21.0 |
21-1 | 绿豆 | 23.4 | 25-2 | 菜豆 | 22.5 |
22-1 | 绿豆 | 26.1 | 27-2 | 菜豆 | 31.7 |
23-1 | 绿豆 | 35.8 | 28-2 | 菜豆 | 34.8 |
3 讨论
随着分子生物学技术的发展,菜豆晕疫病菌的PCR检测方法备受关注,目前报道的特异引物/探针多达14对,但尚无对其特异性和灵敏度进行测试和评价的报道。本研究收集了25株菜豆和绿豆Psph阳性菌株,以及34株假单胞菌属的近似种菌株,用14对引物/探针进行测试,筛选出了最优引物/探针。
菜豆晕疫病菌致病的主要因素是可以产生菜豆毒素(phaseolotoxin),菜豆毒素是一种非寄主专化性毒素,它能对植物和其他微生物产生伤害。菜豆晕疫病菌拥有的菜豆毒素合成基因簇(argK-tox),一方面指导菜豆毒素的生物合成,一方面使寄主增加毒性。argK-tox基因簇只存在于菜豆晕疫病菌、丁香假单胞菌猕猴桃致病变种(Pseudomonas syringae pv. actinidiae)和一株侵染野豌豆的Pseudomonas syringae pv. syringae菌株]。专家普遍认为,能合成毒素基因的Psph菌株(Tox+)才有流行病学意义,因此该DNA区域常常作为PCR检测的靶]。但最新报道表明,超过60%的西班牙Psph菌株不能合成毒素基因(Tox-),也不能与argK-tox靶区引物产生特异性扩增,因此认为这些菌株中argK-tox基因部分或全部缺]。基因分析表明Tox+和Tox-菌株明显分成了两种遗传谱],且Tox+菌株引起的典型症状是病斑圆形、水渍状,周围有绿黄色晕],而Tox-菌株引起水渍状病斑但不产生晕]。本研究收集了目前报道的14对Psph特异引物/探针,检测靶区涵盖了argK-tox和非argK-tox基因区,根据特异性测试筛选出来的引物52B/8F和探针PsF-tox 286P,检测靶区都在argK-tox基因区,对Tox-菌株可能产生假阴性结果,但因本研究未收集到西班牙Psph菌株而暂未得到验证。
Rico]专门针对Tox-菌株设计了引物P3004L/P3004R,与P5.1/P3.1和PHA19/PHA95配合使用来检测Tox+和Tox-菌株。本研究中,来源于菜豆的14株Psph阳性菌株,其中13株经P5.1/P3.1和PHA19/PHA95检测为阳性,同时P3004L/P3004R检测为阴性,但来源于菜豆的LMG2245和来源于绿豆的11株Psph经P5.1/P3.1、PHA19/PHA95和P3004L/P3004R同时检测为阳性,且引物P3004L/P3004R是针对菜豆Psph菌株设计的,目前尚无用于绿豆菌株验证的报道,因此P3004L/P3004R可能只对菜豆Tox-菌株有检测效果,对绿豆菌株没有检测效果。对不同寄主Tox-菌株的特异性检测还需开发更加完善的引物/探针。
本研究实时荧光PCR测试结果表明,探针SSRP_P只能检测来源于菜豆的14株Psph阳性菌株,不能检测其余来源于绿豆的11株Psph,说明探针SSRP_P不能检测绿豆菌株,会造成检测结果假阴性,实际检测中可能造成漏检。Noble]比较了常规PCR、染料法荧光PCR和探针法荧光PCR方法对绿豆晕疫病菌的检测灵敏度,证实了探针法荧光PCR灵敏度最高,这与本研究常规PCR和探针法荧光PCR的灵敏度测试结果一致,但报道中使用的引物P5.1/P3.1特异性较差,可造成假阳性,而探针Psy-cyoII-F/Psy-cyoII-R/Psy-cyoII-pb不是Psph特异探针,可同时检测P. s. pv. syringae等8个致病变种。在口岸实际检测中,样品内部病原菌种类复杂,为准确鉴定有害生物,检测方法的特异性比灵敏度更重要,因此建议使用特异性好的特异引物/探针进行筛选和检测。argK-tox靶区引物和非argK-tox靶区引物检测Psph阳性菌株LMG2245结果均为阳性,其本身是否含有argK-tox基因簇,以及与Tox+和Tox-菌株的序列差异,有待进一步分析。探针SSRP_P的测试结果表明菜豆和绿豆Psph菌株基因存在差异,具体差异也还需要更深一步的研究。
4 结论
本研究收集了25株菜豆和绿豆Psph阳性菌株,以及34株假单胞菌属的近似种菌株,用14对Psph引物/探针进行特异性测试,并对特异性强的引物/探针进行了灵敏度评价,最终筛选出最优引物52B/8F和探针PsF-tox 286P。用常规PCR和探针法实时荧光PCR两种方法对实际样品进行检测,结果表明,引物52B/8F和探针PsF-tox 286P均可用于豆类样品Psph检测。本研究为口岸Psph鉴定提供了一种新的研究方法,也为豆类样品PCR初筛和Psph鉴定提供了一定的技术支撑。
参考文献
[1] Birch R G, Alvarez A M, Patilm S S. A bacterial leaf spot causedin Yam bean by Pseudomonas syringae pv. phaseolicola[J]. Phytopathology, 1981, 71: 1289-1293.
[2] Hunter P J, Taylor J D. Patterns of interaction between isolates of three pathovars of Pseudomonas syringae and accessions of a range of host and non-host legume species[J]. Plant Pathology, 2006, 55: 46-53.
[3] Patel P N, Walker J C. Resistance in Phaseolus to halo blight[J]. Phytopathology, 1965, 55: 889-894.
[4] Taylor J D, Teverson D M, Allen D J, et al. Identification and origin of races of Pseudomonas syringae pv. phaseolicola from Africa and other bean growing areas[J]. Plant Pathology, 1996, 45: 469-478.
[5]陈曦, 张明哲, 林晓佳, 等. 菜豆晕疫病检测技术研究进展[J]. 安徽农业科学, 2013, 41(10): 4365-4367.
[6] Walker J C, Patel P N. Splash dispersal and wind as factors in epidemiology of halo blight of bean[J]. Phytopathology, 1964, 54: 140-148.
[7]魏亚东. 菜豆晕疫病[J]. 天津农林科技, 1994(3): 30-31.
[8]朱振东, 灿星. 绿豆病虫害鉴定与防治手册[M]. 北京: 中国农业科学技术出版社, 2012, 55-62.
[9] Van Schoonhoven A, Voysest O. Common beans in Latin America and Their Constraints[J]. Bean production problems in the tropics, 1989: 33-57.
[10] Prosen D, Hatziloukas E, Schaad N W, et al. Specific detection of Pseudomonas syringae pv. phaseolicola DNA in bean seed by polymerase chain reaction-based amplification of a phaseolotoxin gene region[J]. Phytopathology, 1993, 83(9): 965-970.
[11] Schaad N W, Cheong S S, Tamaki S, et al. A combined biological and enzymatic amplification (BIO-PCR) technique to detect Pseudomonas syringae pv. phaseolicola in bean seed extracts[J]. Phytopathology, 1995, 85: 243-248.
[12] Audy P, Braat C E, Saindon G, et al. A rapid and sensitive PCR-based assay for concurrent detection of bacteria causing common and halo blights in bean seed[J]. Phytopathology, 1996, 86: 361-366.
[13] Mosqueda-Cano G, Herrera-Estrella L. A simple and efficient PCR method for the specific detection of Pseudomonas syringae pv phaseolicola in bean seeds[J]. World Journal of Microbiology and Biotechnology, 1997, 13(4): 463-467.
[14] Marques A S A, Corbière R, Gardan L, et al. Multiphasic approach for the identification of the different classification levels of Pseudomonas savastanoi pv. phaseolicola[J]. European Journal of Plant Pathology, 2000, 106: 715-734.
[15] Rico A, Erdozáin M, Ortiz-Barredo A, et al. Short communication. Detection by multiplex PCR and characterization of nontoxigenic strains of Pseudomonas syringae pv. phaseolicola from different places in Spain[J]. Spanish Journal of Agricultural Research, 2006, 4: 261-267.
[16] Cho J H, Jeong M J, Song M J, et al. Development of PCR Primers to Detect Pseudomonas savastanoi pv. phaseolicola from the Bean Seeds[J]. Research in Plant Disease, 2010, 16(2): 129-135.
[17]杨万风, 刘艳, 刘翔, 等. 进口大豆中菜豆晕疫病菌巢式PCR检测方法的建立[J]. 中国油料作物学报, 2015, 37(1): 113-118.
[18]杨万风, 刘艳, 刘翔, 等. 双重PCR法同步检测菜豆晕疫病菌和萎蔫病菌[J]. 浙江农业学报, 2015, 27(9): 1612-1618.
[19] Schaad NW, Berthier-Schaad Y, Knorr D. A high throughput membrane BIO-PCR technique for ultra-sensitive detection of Pseudomonas syringae pv. Phaseolicola[J]. Plant Pathology, 2007, 56: 1-8.
[20] Cho M S, Jeon Y H, Kang M J, et al. Sensitive and specific detection of phaseolotoxigenic and nontoxigenic strains of Pseudomonas syringae pv. phaseolicola by TaqMan real-time PCR using site-specific recombinase gene sequences[J]. Microbiological research, 2010, 165(7): 565-572.
[21] GBT 28075—2011 萨氏假单胞杆菌菜豆生致病型检疫鉴定方法[S]. 北京: 中国标准出版社, 2011.
[22]周密密, 刘晓宇, 杨静, 等. 日本菜豆种子中菜豆晕疫病菌的检疫鉴定[J]. 植物检疫, 2022, 36(2): 45-50.
[23] Aguilera S, Lopez-lopez K, Nieto Y, et al. Functional characterization of the gene cluster from Pseudomonas syringae pv. phaseolicola NPS3121 involved in synthesis of phaseolotoxin[J]. Journal of Bacterilolgy, 2007, 189: 2834-2843.
[24] de la Fuente-Martı'nez J M, Mosqueda-Cano G, AlvarezMorales A, et al. Expression of a bacterial phaseolotoxin-resistant ornithyl transcarbamylase in transgenic tobacco confers resistance to Pseudomonas syringae pv. Phaseolicola[J]. Biotechnology, 1992, 10: 905-909.
[25] Tamura K, Takikawa Y, Tsuyumu S, et al. Characterization of the toxin produced by Pseudomonas syringae pv. actinidiae, the causal bacterium of kiwifruit canker[J]. Japanese Journal of Phytopathology, 1989, 55(4): 437-444.
[26] Tourte C, Manceau C. A strain of Pseudomonas syringae which does not belong to pathovar phaseolicola produces phaseolotoxin[J]. European Journal of Plant Pathology, 1995, 101: 483-490.
[27] Rico A, López R, Asensio C, et al. Nontoxigenic strains of Pseudomonas syringae pv. phaseolicola are a main cause of halo blight of beans in Spain and escape current detection methods[J]. Phytopathology, 2003, 93: 1553-1559.
[28] Oguiza J A, Rico A, Rivas L A, et al. Pseudomonas syringae pv. phaseolicola can be separated into two genetic lineages distinguished by the possession of the phaseolotoxin biosynthetic cluster[J]. Microbiology, 2004, 150: 473-482.
[29] Taylor J D, Dudley C L, GRAY L P. Studies of halo-blight seed infection and disease transmission in dwarf beans[J]. Annals of Applied Biology, 1979, 93(3): 267-277.
[30] Patil S, Hayward A, Emmons R. An ultraviolet-induced nontoxigenic mutant of Pseudomonas phaseolicola of altered pathogenicity[J]. Phytopathology, 1974, 64: 590-595.
[31] González A I, De la Vega M P, Ruiz M L, et al. Analysis of the argKtox gene cluster in nontoxigenic strains of Pseudomonas syringae pv. phaseolicola[J]. Applied and Environmental Microbiology, 2003, 69: 4979-4982.
[32] Noble T J, Williams B, Douglas C A, et al. Evaluating molecular diagnostic techniques for seed detection of Pseudomonas savastanoi pv. phaseolicola, causal agent of halo blight disease in mungbean (Vigna radiata) [J]. Australasian Plant Pathology, 2022, 51(4): 453-459.
基金项目:广东省口岸安全智能化检测重点实验室(2023B1212010011)
第一作者:牟桂萍(1987—),女,汉族,山东邹平人,农艺师,主要从事植物病原细菌检疫鉴定研究工作,E-mail: mgping1234@163.com
通信作者:易建平(1969—),男,汉族,四川乐山人,研究员,主要从事植物病原细菌检疫鉴定研究工作,E-mail: yjp16@126.com
1. 广东省口岸安全智能化检测重点实验室 广州 510700
2. 黄埔海关技术中心 广州 510700
3. 上海海关动植物与食品检验检疫技术中心 上海 200135
1. Guangdong Provincial Key Laboratory of Intelligent Port Security Inspection, Guangzhou 510700
2. Technology Center of Huangpu Customs, Guangzhou 510700
3. Animal, Plant and Food Inspection and Quarantine Technology Center of Shanghai Customs, Shanghai 200135
表1 引物/探针信息
Table 1 Information of primers and probes
方法 | 序号 | 引物名称 | 序列 (5' -3' ) | 产物 (bp) | 基因名称 | Tm值(℃) | 参考文献 |
常规PCR | 1 | HM6 | CGTGTCCGTGGATAAAAGC | 1900 | argK | 60 | |
HM13 | GTTGAATTTCACTACCCG | ||||||
2 | P5.1 | AGCTTCTCCTCAAAACACCTGC | 500 | argK-tox | 58 | ||
P3.1 | TGTTCGCCAGAGGCAGTCATG | ||||||
3 | P5.2 | TCGAACATCAATCTGCCAGCCA | 450 | argK-tox | 58 | ||
P3.2 | GGCTTTTATTATTGCCGTGGGC | ||||||
4 | HB14F | CAACTCCGACACCAGCGACCGAGC | 1413 | argK | 65 | ||
HB14R | CCGGTCTGCTCGACATCGTGCCAC | ||||||
5 | 62a | CAATGAAGATTACAAGCCTG | 1000 | argK | 60 | ||
63a | GCTAGCTATCAGGGGACGAC | ||||||
6 | PHA19 | CGTCTGTAACCAGTTGATCC | 437 | amtA | 52 | ||
PHA95 | GAATCCTTGAATGCGAAGGC | ||||||
7 | P3004L | CTGTCTGGCAGCCACTACAAAG | 243 | AP-PCR sequence | 58 | ||
P3004R | GGCTGCAAATTGTGGGATTT | ||||||
8 | psp-JH-F | GCATCAGCATTTCGACCTTT | 513 | argK | 58 | ||
psp-JH-R | ATGGGCGAGAAACATTCAAC | ||||||
常规PCR | 9 | psp-JH-F-ne | GCATCAGCATTTCGACCTTT | 169 | argK | 58 | |
psp-JH-R-ne | CAATCAAGTGGCACGATGTC | ||||||
10 | 52B | GTTTCGCTCGGGAGTCTG | 419 | argK | 60 | ||
8F | ATGAAGGAGCGCACTTTCA | ||||||
11 | 24B | GTTTCGGCCGTCACCAAC | 224 | argK | 60 | ||
24F | AGGATTTTGCCCGTGTTGT | ||||||
12 | PSPF1 | GACGACTGTCTCCAGCATCAG | 533 | argK | 60 | ||
PSPR2 | AAGAATTCGGGCGCATTGA | ||||||
实时荧光PCR | 13 | PsF-tox 188 | GGGGTGGGACGTGTTAT | 369 | tox-argK | 60 | |
PsR-tox 557 | GAACATCAATCTGCCAGCC | ||||||
PsF-tox 286P | FAM-ACCATCCGAATGCCAGTAATGCC-TAMRA | ||||||
14 | SSRP_F | GACGTCCCGCGAATAGCAATAATC | 183 | site-specific recombinase gene | 61 | ||
SSRP_R | CAACGCCGGCGCAATGTCG | ||||||
SSRP_P | FAM-TGACGTGACACTCGCCGAGCTGCA-BHQ1 |
表1(续)
表2 试验菌株和PCR检测结果
Table 2 Test strains and PCR results
名称 | 菌株编号 | 寄主 | 年份 | 引物序号 | |||||||||||||
1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | 14 | ||||
Pseudomonas savastanoi pv. phaseolicola | LMG2245a | 菜豆 | 未知 | + | + | + | + | + | + | + | + | + | + | + | + | + | + |
ATCC11355b | 菜豆 | 未知 | + | + | + | + | + | + | - | + | + | + | + | + | + | + | |
1199-1c | 菜豆 | 2021 | + | + | + | + | + | + | - | + | + | + | + | + | + | + | |
2199-4d | 菜豆 | 2021 | + | + | + | + | + | + | - | + | + | + | + | + | + | + | |
3521-1 | 菜豆 | 2019 | + | + | + | + | + | + | - | + | + | + | + | + | + | + | |
424-1e | 菜豆 | 2017 | + | + | + | + | + | + | - | + | + | + | + | + | + | + | |
5824-1 | 菜豆 | 2015 | + | + | + | + | + | + | - | + | + | + | + | + | + | + | |
8253f | 菜豆 | 2015 | + | + | + | + | + | + | - | + | + | + | + | + | + | + | |
7185-1 | 菜豆 | 2022 | + | + | + | + | + | + | - | + | + | + | + | + | + | + | |
7186-3 | 菜豆 | 2022 | + | + | + | + | + | + | - | + | + | + | + | + | + | + | |
7253-1 | 菜豆 | 2022 | + | + | + | + | + | + | - | + | + | + | + | + | + | + | |
7254-1 | 菜豆 | 2022 | + | + | + | + | + | + | - | + | + | + | + | + | + | + | |
7281-1 | 菜豆 | 2022 | + | + | + | + | + | + | - | + | + | + | + | + | + | + | |
7282-1 | 菜豆 | 2022 | + | + | + | + | + | + | - | + | + | + | + | + | + | + | |
7181-1 | 绿豆 | 2022 | + | + | + | + | + | + | + | + | + | + | + | + | + | - | |
7182-1 | 绿豆 | 2022 | + | + | + | + | + | + | + | + | + | + | + | + | + | - | |
Pseudomonas syringae pv. phaseolicola | 7183-1 | 绿豆 | 2022 | + | + | + | + | + | + | + | + | + | + | + | + | + | - |
7184-1 | 绿豆 | 2022 | + | + | + | + | + | + | + | + | + | + | + | + | + | - | |
7191-1 | 绿豆 | 2022 | + | + | + | + | + | + | + | + | + | + | + | + | + | - | |
7256-1 | 绿豆 | 2022 | + | + | + | + | + | + | + | + | + | + | + | + | + | - | |
7257-1 | 绿豆 | 2022 | + | + | + | + | + | + | + | + | + | + | + | + | + | - | |
7258-1 | 绿豆 | 2022 | + | + | + | + | + | + | + | + | + | + | + | + | + | - | |
7259-1 | 绿豆 | 2022 | + | + | + | + | + | + | + | + | + | + | + | + | + | - | |
72510-1 | 绿豆 | 2022 | + | + | + | + | + | + | + | + | + | + | + | + | + | - | |
8765-1 | 绿豆 | 2022 | + | + | + | + | + | + | + | + | + | + | + | + | + | - | |
P. s. pv. actinidiae | Psa01g | 猕猴桃 | 2011 | - | - | +L | - | - | - | + | - | - | - | - | - | - | - |
Psa02g | 猕猴桃 | 2011 | - | - | +L | - | - | - | + | - | - | - | - | - | - | - | |
Psa03g | 猕猴桃 | 2011 | - | - | +L | - | - | - | + | - | - | - | - | - | - | - | |
5023h | 猕猴桃 | 2022 | - | - | +L | - | - | +L | + | - | - | - | - | - | - | - | |
P. s. pv. pisi type I | 5337-2 | 豌豆 | 2017 | - | - | - | - | - | +L | + | - | - | - | +L | - | - | - |
P. s. pv. pisi type II | 6339-2 | 豌豆 | 2020 | - | - | - | - | - | +L | + | +L | - | - | - | - | - | - |
P. s. pv. syringae | 0796-3 | 樱桃 | 2018 | - | - | - | - | - | - | + | +L | +L | - | +L | - | - | - |
0796-4 | 樱桃 | 2018 | - | - | +L | +L | - | - | + | +L | +L | - | - | - | - | - | |
0796-9 | 樱桃 | 2018 | - | - | +L | - | - | - | + | +L | - | - | - | - | - | - | |
918T-4i | 大豆 | 2012 | - | - | - | - | - | - | + | - | - | - | - | - | - | - | |
P. s. pv. tomato | 7301-1 | 番茄 | 2015 | - | - | - | +L | - | - | + | - | +L | - | - | - | - | - |
7281 | 番茄 | 2015 | - | - | - | +L | - | - | + | - | - | - | - | - | - | - | |
P. s. pv. tabaci | T5j | 未知 | 2008 | - | - | - | - | - | - | + | - | - | - | +L | +L | - | - |
P. s. pv. glycinea | 918P-1 i | 大豆 | 2012 | - | - | - | - | - | - | + | - | - | - | - | +L | - | - |
psg363k | 大豆 | 2012 | - | - | - | - | - | - | + | - | - | - | - | +L | - | - | |
MT-1 i | 大豆 | 2011 | - | - | - | - | - | - | + | - | - | - | - | +L | - | - | |
P. s. pv. maculicola | 9951-9 | 油菜 | 2021 | - | - | - | - | - | - | + | - | - | - | - | - | - | - |
2475-1 | 油菜 | 2021 | - | - | - | - | - | - | + | - | - | - | - | - | - | - | |
P. s. pv. morsprunorum | 4458-3 i | 樱桃 | 2012 | - | - | - | - | - | - | + | - | +L | - | - | +L | - | - |
6158-4 | 樱桃 | 2018 | - | - | - | - | - | - | + | - | - | - | - | +L | - | - | |
P. s. pv. persicae | NCPPB3687l | 李 | 1990 | - | - | - | - | - | - | + | - | - | - | +L | - | - | - |
P. s. pv. lachymas | NCPPB 540m | 黄瓜 | 1958 | - | - | - | - | - | - | + | +L | +L | - | - | +L | - | - |
Ps18 | 未知 | 未知 | - | - | - | - | - | - | + | +L | - | - | - | +L | - | - | |
P. sp. | 3993-2 | 豌豆 | 2021 | - | - | + | - | - | + | + | - | - | - | - | - | - | - |
4593-4 | 菜薹 | 2020 | - | - | - | - | - | - | + | - | - | - | +L | +L | - | - | |
4593-6 | 菜薹 | 2020 | - | - | - | - | - | - | + | - | - | - | - | +L | - | - | |
P. cichorii | ZSXF02 | 未知 | 2020 | - | +L | +L | - | - | - | + | - | - | - | - | - | - | - |
ND04A | 未知 | 2020 | - | +L | +L | - | - | - | + | - | - | - | - | - | - | - | |
P. viridiflava | 2135b | 未知 | 2021 | - | +L | - | - | - | +L | + | +L | +L | - | +L | +L | - | - |
525-1 | 芸薹 | 2021 | - | +L | - | - | - | +L | + | +L | +L | - | +L | +L | - | - | |
525-2 | 芸薹 | 2021 | - | +L | - | - | - | +L | + | +L | +L | - | +L | +L | - | - | |
0652-2 | 豌豆 | 2022 | - | +L | - | - | - | +L | + | - | - | - | +L | +L | - | - | |
NCPPB2254 | 桃 | 1972 | - | +L | - | - | - | +L | + | - | - | - | +L | +L | - | - | |
P. cannabina pv. alisalensis | NCPPB1820 l | 萝卜 | 1966 | - | - | +L | - | - | - | + | - | - | - | - | - | - | - |
注:①+: 阳性; -: 阴性; +L: 弱阳性; ②1: HM6/HM13; 2: P5.1/P3.1; 3: P5.2/P3.2; 4: HB14F/HB14R; 5: 62a/63a; 6: PHA19/PHA95; 7: P3004L/P3004R; 8: psp-JH-F/psp-JH-R; 9: psp-JH-F-ne/psp-JH-R-ne; 10: 52B/8F; 11: 24B/24F; 12: PSPF1/PSPR2; 13: PsF-tox 188/PsR-tox 557/PsF-tox 286P; 14: SSRP_F/SSRP_R/SSRP_P ; ③a.李斌, 浙江大学; b.刘鹏, 天津海关; c.王溪桥, 兰州海关; d.吴翠萍, 南京海关; e.王翀, 乌鲁木齐海关; f.文朝慧, 兰州海关; g.胡白石, 南京农业大学; h. 邵宝林, 成都海关; i.叶露飞, 舟山海关; j.姬广海, 云南农业大学; k.冯建军, 深圳海关; l.朱金国, 长沙海关; m.赵文军, 中国检验检疫科学研究院.
表2(续)
表2(续)
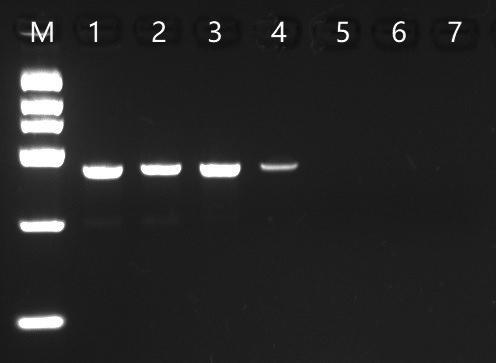
M: DL2000 Marker; DNA添加量(1—7) 依次为50 ng、5 ng、500 pg、50 pg、5 pg、500 fg、50 fg.
图1 常规PCR引物52B/8F灵敏度测试
Fig.1 Sensitivity test of PCR assay using primer 52B/8F
DNA添加量 (1—7) 依次为50 ng、5 ng、500 pg、50 pg、5 pg、500 fg、50 fg.
图2 实时荧光PCR探针PsF-tox 286P灵敏度测试
Fig.2 Sensitivity test of real-time PCR assay using probe
PsF-tox 286P
PC: 阳性对照; NC: 阴性对照; CK: 空白对照
图3 常规PCR引物52B/8F测试样品
Fig.3 Sample test of PCR assay using primer 52B/8F